DeepETD and EMTDD
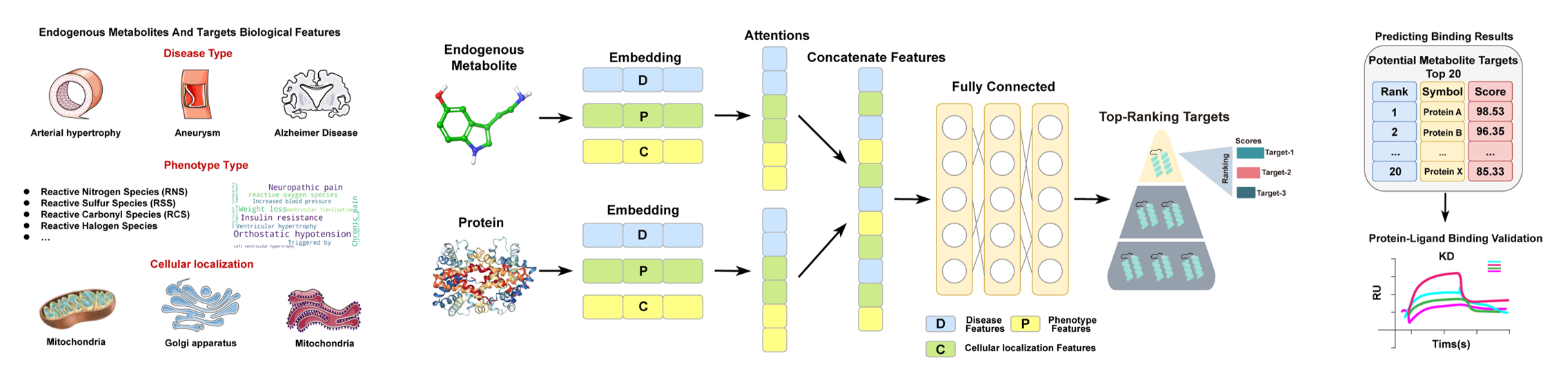
Metabolites are involved in almost all fundamental biological processes. The identification of their protein targets is crucial for elucidating non-canonical signaling roles and evaluating their therapeutic potential. In recent years, due to the continuous development of chemical proteomics, atypical phenotypic functions of classic metabolites have been continuously discovered. However, targets discovery of endogenous metabolites for their pathology-related functions is still progressing slowly. To accelerate the identification of disease-associated targets for endogenous metabolites, we propose a novel hypothesis: endogenous metabolites and their molecular targets are expected to possess similar disease correlations. Following this hypothesis, here we report the development of a novel deep-learning model named DeepETD, integrating bioinformatic data and introducing an attention mechanism to predict the functional targets of specific metabolite phenotypes. Using this model, we constructed a publicly accessible database named EMTDD containing potential targets for 3,382 common human endogenous metabolites.
Citation 1
DeepETD: A computational tool for endogenous metabolite target discovery based on disease correlation mapping
PMID : XXXXX
Citation 2
Norepinephrine aggravates atherosclerosis by inhibiting PRDX1: Evidence from a new deep-learning model
PMID : XXXXX